Puma plotting tutorial
Introduction
In this tutorial you will learn how to use puma, the Plotting UMami Api. The idea behind puma is to provide a plotting API that is easy to use but at the same time highly configurable. This means that the user has full control over the plot while things like uncertainties, labels, ratio panels can be added easily.
You can find the puma documentation here.
puma is based on matplotlib and helps you to produce most of the types of plots that are commonly used in flavour tagging like the ones shown below:


In this tutorial you will learn how to:
A small introduction to the different metrics used in flavour tagging.
Plot histograms with
puma, where you will produce plots of both jet- and track-variables.Plot ROC curves of two taggers, with ratio panels that compare the curves.
Plot the efficiency/rejection of a tagger as a function of pt.
The tutorial is meant to be followed in a self-guided manner. You will be prompted to do certain tasks by telling you what the desired outcome will be, without telling you how to do it. In case you are stuck, you can click on the "hint" toggle box to get a hint. If you tried for more than 10 min at a problem, feel free to toggle also the solution with a worked example.
Setting Up Everything You Need
A pleasant way to work through the tutorial is using a jupyter notebook environment.
Jupyter Lab on DESY NAF: http://naf-jhub.desy.de
Documentation: https://confluence.desy.de/display/IS/Jupyter+on+NAF
Display images in jupyter notebook
The scripts in the tutorial save the images as png files. You can display them in the notebook with this piece of code in a python cell:
from IPython.display import Image
Image("path_to_image")
Alternatively, you can use markdown syntax in a markdown cell:

For the tutorial, you will need puma. The puma package can be easily installed via the command
If you are using jupyterlab on DESY NAF, then using the ftag Kernel will provide you already with puma installed, so you don't need to install it yourself.
For this tutorial, two .h5 files were prepared, one sample of top quark pairs and one Z' file, providing high-pt jets originating from a hypothetical heavy spin-1 particle.
They are located here at the DESY NAF computing infrastructure:
Tutorial tasks
The tasks are divided in several sub-tasks. You don't have to do all of them in case you are more interested in the other tasks. However, the sub-tasks depend on each other, so you should finish a subtask before proceeding to the next one (also the solutions assume that you already have the previous subtasks completed).
Task 1: Flavour tagging metrics
To get started with the plotting, we will first have a quick look into different metrics used in flavour tagging to evaluate the performance.
Task 1.1: Generating dummy data
The typical output of our ML-based taggers, like DIPS or DL1d, are 3 scores indicating the probabilities of being a light-flavour jet, a c-jet and a b-jet. For example the scores [0.1, 0.3, 0.6] would indicate that we have most probably a b-jet while [0.7, 0.2, 0.1] would indicate a light-flavour jet.
Even though we always have MC simulation, it is sometimes useful to have dummy data, for instance if we want to test certain plotting features etc. (and to understand the actual underlying distributions).
Now, it is up to you. Generate a dummy multi-class output of a neural network.
Hint: Where can I find such a function?
You can have a look at the puma documentation and search in the API reference.
Hint: Which exact function?
the puma.utils.generate module contains the desired functions
Task 1.2: Defining Working points - Likelihood ratio
Since we are not (yet) able to calibrate the entire spectrum of the different multi-class outputs, we need to define so-called working points (or operating points). In the case of ATLAS, we have four different b-tagging working points (WPs) which are defined covering various needs of the physics analyses. The efficiency of a specific flavour j (b, c or light) is defined as
where Npassj(D>Tf) are the number of jets of flavour j passing the cut Tf on the tagger discriminant D and Ntotalj are all jets of flavour j before the cut.
The final output score is calculated from the multi-class output and results for the b-tagging discriminant into the log-likelihood
with pb, pc, pl being the probabilities for the jet to be a b-jet, c-jet or light-flavour jet, respectively. The c-jet fraction fc allows to tune how much emphasis is given to the c-jet or to the light-flavour performance. While the c-jet rejection increases as a function of fc, the light-flavour jet rejection decreases. This parameter has to be tuned separately for each tagger and depends on the needs of the physics analyses.
The advantage of the multi-class output is that this tuning is possible after the training and the c-jet fraction in the training sample does not have to be adapted. Another advantage of the multi-class output is that one can by changing the log-likelihood to
perform c-tagging without the need of retraining the tagger. Here fb is now the b-jet fraction.
Define a function which calculates the log-likelihood, when giving it the 3 scores and the fc value as input.
Solution
You can find the solution in the histogram example of the puma documentation.
Using the dummy data from task 1.1, calculate the log-likelihood with fc =0.018 and retrieve the working point cut value for 70% b-jet efficiency.
Hint: Where to get the labels from?
The labels from task 0.1 have the same values as the HadronConeExclTruthLabelID.
The standard labelling is provided via the HadronConeExclTruthLabelID variable, which is defined as follows:
For matching all weakly-decaying b-/c-hadrons and tau-leptons with a transverse momentum larger than 5 GeV and all calorimeter (track) jets with an uncalibrated transverse momentum of at least 0 (5) GeV are found. A hadron/tau-lepton is matched with a jet if the angular distance DeltaR is smaller than 0.3. In case of multiple jets laying in this cone the closest one is chosen. Afterwards all hadrons/tau-leptons that are daughter particles of another hadron/tau-lepton are removed. The remaining ones define the label for the jet:
light jets: 0
c-jets: 4
b-jets: 5
tau-jets: 15
Hint: Which function to use?
You can have a look at the percentile function from numpy. Be aware from which site we need to integrate! And the apply_along_axis function to evaluate an entire array.
Solution
You can also have a look at the metrics function in puma , where this code is also being used.
Task 1.3: Performance metrics - efficiency and rejection
To quantify the performance of a tagger at a given working point, the background rejection is a good measure. The rejection is simply the inverse of the efficiency 1/εi.
Calculate the light-flavour jet and c-jet rejection for the 70% working point from task 0.2.
Solution
Alternatively, all this functionality is also provided by puma.metrics. In that case this would simplify to
Starting from these metrics, we can plot for instance:
ROC curves: which show the background rejection as function of the b-jet efficency
Efficiency vs pT: where one fixes a working point and calculates the background rejection in bins of pT
Task 2: Histogram plots
Task 2.1: Loading the h5 file
Before starting with the different plotting exercises, you have to load the h5 file that was prepared for this tutorial. The expected outcome of this is that you have access to the jet variables as well as to the track variables. You can put the jet variables in a pandas.DataFrame in case you feel more comfortable with that, but this will not be possible for the tracks, since the tracks_loose dataset in the h5 file has an extra dimension for the tracks (for each jet we store the information of up to 40 tracks).
Write a little python script that loads the jet and track variables.
Have a look at how large the dataset is, and what shape the loaded arrays have.
For the following tasks you can re-use the code that loads the h5 file or just extend your python script from this task.
Remember, that the ttbar h5 file is located here on NAF:
Hint: how can I load a h5 file with python?
You can find the documentation of h5py here.
Task 2.2: Plotting the pT distribution for jets of different flavours
As a next step, you will produce a histogram plot that shows the pT distribution of light-flavour jets, c-jets and b-jets.
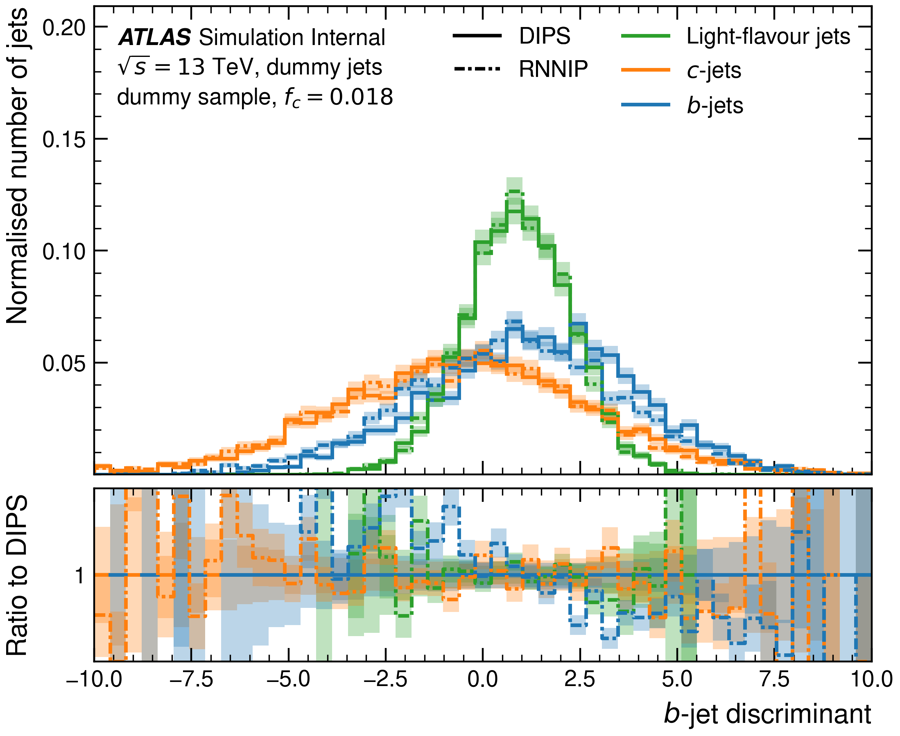
Task 2.3: Plot the b-jets probability output of two different taggers
In this task you will plot the b-jets probability of two different taggers - RNNIP and DIPS.
Create the histogram plot (similar to the one from the previous task) and the different histograms. If you plot this for light-flavour jets, c-jets and b-jets, you should have 6 histograms.
Make sure that you use a different line-style for the histograms of you second tagger.
Add a ratio panel to the plot
Make your plot look pretty. Have a look at the arguments that are supported by
puma.PlotObject.
Hint 1: Histogram and HistogramPlot objects
After you defined your HistogramPlot object, you can start adding lines to it. This lines are the Histogram objects you need to define.
Hint 3: Ratio Panel
The ratio for the ratio panel is calculated in this case between the same flavours. But you need to tell the plot which of the Histogram objects is the reference. Try to look up the reference option in the add() function.
Task 2.4: Plot a track variable of your choice
In this task you are asked to make a histogram plot of a track variable. This is slightly more tricky, since the array that you load from the h5 file has a different shape compared to the array storing the jet information. In addition to that, many entries might be filled with nan values, which is challenging here and there.
Choose a track variable that you want to plot.
Create a histogram plot (maybe again for multiple flavours, but that is up to you).
Hint 1: NaN-Values in binning
If you encounter an issue with NaN values in the binning, you need to set the bins_range correctly, because with NaN values it cannot be calculated automatically.
Hint 2: Difference in Shape
Due to the dimensionality of tracks, you need to get rid of one of the dimensions. Try the .flatten() option of numpy.ndarray.
Task 3: ROC plots
In this task, you will plot a ROC comparison for the two taggers RNNIP and DIPS.
Task 3.1: Calculate the rejections as a function of the b-jets efficiency
Before you can actually plot the ROC curves, you have to calculate the light-flavour and c-jets rejection for a range of b-jets efficiencies.
Define a function that calculates the b-jets discriminant from the tagger output.
Calculate the light-flavour jets rejection as a function of the b-jets efficiency.
Task 3.2:
Plot the light-flavour jets rejection as a function of the $b$-jets efficiency. Use
n_ratio_panels=1to also get the ratio of the two rejection curves.
Hint 1: How do I initialise a ROC curve plot?
Plotting ROC curves with puma is similar to plotting histograms. The main difference is that you are using the puma.RocPlot and puma.Roc classes. Search the puma docs for "roc" to have a look at an example and the API reference.
Hint 2: I initialised the plot and added the ROC curves - is there anything else to do?
For ROC curves you also have to define the class which is drawn in the ratio panel. The method you need to use here is RocPlot.set_ratio_class().
Task 3.3: Add the c-rejection to your plot
Repeat the calculation of the rejection for c-jets
Add the corresponding ROC curves to the plot. Don't forget to increase
n_ratio_panelsof yourpuma.RocPlot.
Hint 1: How can I modify the legend that states which linestyle corresponds to the different rejection classes
You can add labels for the different rejection classes using the RocPlot.set_leg_rej_labels() method.
Task 4: pT vs. efficiency
In this task, you will plot both the b-jets efficiency and the light-flavour jets rejection for specific bins of pT.
Task 4.1: Calculate the discriminant values
Just like you did in Task 3.1, calculate the discriminant scores for RNNIP and DIPS. You can reuse the code from task 3.1. If you are putting everything in one python script you can just reuse the values that are already calculated.
Task 4.2: Create a pT vs. b-efficiency plot
For a fixed inclusive b-efficiency, you plot the b-efficiency for different bins of pT.
Task 4.3: Create a pT vs. light-flavour jets rejection plot
Last updated